Wafer scanning process traveler: Difference between revisions
No edit summary |
→Wafers to Use: updated surfscan instructions. |
||
| (119 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
<small>''[[Biljana Stamenic]], NanoFab Staff, April 2020''</small> |
|||
The wafers used for process calibration are ordered from SVM. These are 4 inch silicon wafers with low particle count (LPD= light particle detection<100). The box with 25 wafers is stored in the cleanroom and used only for process calibration. Wafers are handled very carefully, because with every loading and unloading some particles will be added. |
|||
Below is the procedure we use for calibrating particle counts on various machines, primarily depositions. |
|||
==Wafers to Use== |
|||
=== Scanning procedure === |
|||
The wafers for process calibration are ordered from a wafer supplier that sells high grade, quality wafers. These are 4-inch Si wafers, with low particle count (Particles: <=10 @ >=0.3 microns). Wafers should be handled carefully, because every loading and unloading will add some particulates. |
|||
Regular Prime grade wafers also work well, but note that initial pre-process particle measurement may be higher - this is ok as long as you measure ''before'' the process step, and subtract that value from the ''after'' process value. If your ''before'' measurement shows high (eg. >200 particles), do not use that wafer for particle checks, discard it and find a better wafer. |
|||
====Scanning procedure before process calibration==== |
|||
# Log in (access code is boss) |
|||
Surfscan 6200 (located in bay5) is used for scanning wafers. There are two standard recipes that could be used for scanning wafers: UCSBTEST1 (for scanning 8” wafers) and UCSBTEST2 (for scanning 4"wafers). The recipe UCSBTEST2 is used as a template to create other two standard recipes: "UCSB Gain4" (measuring small particles 0.16-1.6um) and "UCSB Gain2" (measuring larger particles 1.6-28um). |
|||
# Load desired number of Si wafers to the carrier that says " SURFSCAN" . |
|||
# Place the carrier with wafers to the right indexer. |
|||
We have cassettes only for 4", 6" and 8" wafers. If you plan to scan wafers that have different size, you need to provide your own cassette and calibrate it. If you need to scan wafer pieces, you need to develop your own procedure how to scan it. |
|||
# Select in the menu option "CAS" (this will read all wafers that are in the carrier) |
|||
# Load one of standard recipes |
|||
==Scanning procedure for 4" Si wafers== |
|||
# Select one of the recipes: |
|||
The '''right-cassette/indexer''' is calibrated for 4-inch and 6-inch Si wafers. The '''left-cassette/indexer''' is available for 8inch Si wafers (not calibrated). |
|||
#* UCSB Gain4 ( measuring small particles 0.16-2.8um) |
|||
#* UCSB Gain2 ( measuring larger particles 2.8-28um) |
|||
===Scan before process step=== |
|||
# Select one of the recipe, modify it if needed, default the bin |
|||
# Select the wafer you want to scan |
|||
#Log in (access code is boss) |
|||
# Press SCAN |
|||
#Load wafers (face up) to the cassette that says " 4inch Surfscan" |
|||
# Wait until scan is finished |
|||
#Place the cassette with wafers to the right indexer |
|||
# Record: LPD, particles #1, particles #8, haze |
|||
#In the menu option select "CASS" (this will read all wafers that are loaded into the cassette) |
|||
# Take a picture of the scan (by phone) |
|||
#Go to "Recipes" |
|||
# Select "Home" so indexer moves up to loading/unloading positon |
|||
#Select one of the recipes: |
|||
# Wafers are ready for process calibration |
|||
#*UCSB G4 0.16-1.6 (measuring small particles 0.16-1.6um) |
|||
====Scanning procedure after process calibration==== |
|||
#*UCSB G2 1.6-28.0 (measuring larger particles 1.6-28um) |
|||
# Log in (access code is boss) |
|||
#**Recipe "UCSBTEST2" is used as a template to create recipes UCSB Gain2 and UCSB Gain4 |
|||
# Load treated Si wafers to the carrier that says " SURFSCAN" . |
|||
#View the recipe and modify it if needed (if recipe is modified, select bin split/default to save changes) |
|||
# Place the carrier with wafers to the right indexer. |
|||
# |
#Select the wafer in the catalog box you want to scan, and press START (it would scan only one wafer) |
||
#Select AUTO option If you want to scan all wafers in the catalog box, followed by START |
|||
# Load one of standard recipes |
|||
#Wait until scan is finished |
|||
# Select one of the recipes : |
|||
#Record: |
|||
#* UCSB Gain4 ( measuring small particles 0.16-2.8um) |
|||
#*'''LPD Cnt''' (sum of all particles #1- #8), particles #1(0.160-0.213)um, particles #8(1.20-1.60)um |
|||
#* UCSB Gain2 ( measuring larger particles 2.8-28um) |
|||
#*'''Haze''' (region(%), average(ppm), peak(ppm)) |
|||
# Select one of the recipe, modify it if needed, default the bin |
|||
#Take a picture of the scan (by phone) |
|||
# Select the wafer you want to scan |
|||
#Select "Home" so indexer moves up to loading/unloading position |
|||
# Press SCAN |
|||
#Unload wafers |
|||
# Wait until scan is finished |
|||
#Log out |
|||
# Record: LPD, particles #1, particles #8, haze |
|||
# Take a picture of the scan (by phone) |
|||
====Data Recording==== |
|||
# Select "Home" so indexer moves up to loading/unloading positon |
|||
# Unload the wafers |
|||
*Run the above process TWICE: once for recipe "Gain 2" (sizes 1.6µm to 28µm) and another for recipe "Gain 4" (sizes 0.160µm to 1.6µm) |
|||
*Record the following: |
|||
**"total particle count", |
|||
**"Haze %" and |
|||
**<u>Take photos of the results screen</u> showing all binning and wafer particle distribution. See example photos below. |
|||
[[File:Surfscan G4 before example - P1 230317E7G4 before.jpg|alt=Photo of Surfscan screen showing low particles on wafer.|none|thumb|381x381px|Typical "before" process step Surfscan measurement. A std. Prime Grade Silicon wafer may have 50-100 particles out of the cassette. Note the tweezer pickup location, on the bottom in this photo.]] |
|||
===Perform process step=== |
|||
*Run the deposition or other process on your wafer. |
|||
*Be careful of adding particles during handling - eg. don't leave wafer out on table unnecessarily, keep it in the case and do not agitate the wafer in the case. |
|||
===Scan after process calibration=== |
|||
*Repeat the same procedure above for scanning. |
|||
*Subtract pre-scan particle counts, to remove pre-process particles that were already on the wafer. |
|||
===Example Surfscan photos=== |
|||
====Low particle example==== |
|||
Here are two example photos for UCSB Gain4, and UCSB Gain2 recipes, showing acceptable particle levels: |
|||
[[File:UCSBTEST2 .png|none|thumb|422x422px|Recipe UCSB Gain4 for smaller particulates (0.160µm to 1.6µm)]] |
|||
[[File:UCSBTEST1.png|none|thumb|425x425px|Recipe UCSB Gain2 for larger particulates (1.6µm to 28µm)]] |
|||
====High Particle Example==== |
|||
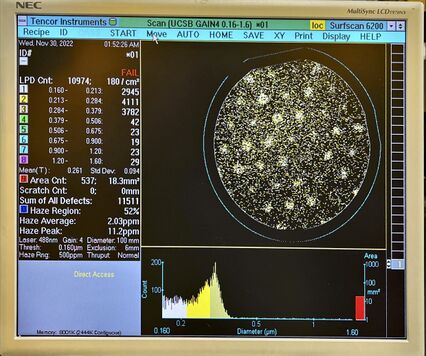
The following shows examples of high particle levels detected ''after'' a deposition process step. |
|||
[[File:Surfscan P1 SiN 221208Z5G4 after.jpg|alt=Photo of surfscan showing high particle count nd hexagonal grid pattern in particle concentrations.|none|thumb|426x426px|Example of high particles detected after deposition step. Gas inlet showerhead pattern is seen, suggesting the source of the particles.]] |
|||
<br /> |
|||
==Plotting Process Control Data== |
|||
Make an Excel spreadsheet. Enter information for Gain4 and Gain2 (particles#1, particles#8, LPD, haze) before and after deposition. For each gain calculate Delta (LPDad - LPDbd). Make a plot (Particulates vs. Date). |
|||
Example : https://docs.google.com/spreadsheets/d/1ct-SSCRxf81W0jupCWzh81n0DBwPv6mwKFMzuWu_0TI/edit#gid=1017053260 |
|||
[[File:PECVD1 particulates in 2019.jpg|none|thumb|643x643px]] |
|||
==Surfscan additional information== |
|||
===Introduction=== |
|||
Surfscan 6200 locates, sizes, and counts defects in semiconductor substrate material down to 0.157um at a 95% capture rate and down to 0.09um at an 80% capture rate. The instrument uses laser beam scanning for detecting defect contamination and displays scan results in color-coded wafer maps, histograms, and summaries. The instrument scans wafers contained in a cassette. Cassettes can be loaded onto the left indexer, right indexer, or both indexers. Cassette configuration can be ( R only, L only, Right to left, Left to right, Sort wafers). When manually aligning wafers, one of the flats of each wafer should face the front of the cassette. When using a mechanical aligner, the flats should face the rear of the cassette. If wafers get scratched, damaged or mishandled immediately perform cassette calibration. The instrument communicates with an operator using the Microsoft Windows. To use an application, choose menus or commands displayed in the menu bar, or use keypad commands. The system menu is an icon-based menu. Double click on the application icon to start an application. In the Scan window , the menu bar displays: ID, CASS, AUTO, HOME, SAVE, and PRINT commands. |
|||
===Basic skills=== |
|||
This chapter describes instrument controls, including keypad, mouse, floppy drive, and cassette indexer. This chapter also describes how to load a cassette and introduces you to basic program operation using the mouse or keypad. |
|||
===Getting started=== |
|||
This chapter provides basic startup procedures for the instrument, including: (Powering up the instrument , Logging on to the instrument , Starting Scan , Quitting from an application, Logging off the system, Setting up the system for first use, Using the DOS shell (advanced use), Monitoring disk and database space) |
|||
Log in, access code is boss. |
|||
Double click on the icon of the application to start application. |
|||
Log off the system, to exit the current application. |
|||
The Tencor Instrument banner at the top of the screen contains a disk status. If it is green the database space is available. If it is yellow, free disk or data space is getting low, so it needs to be increased free space. It is red, there is no free space. |
|||
===Scanning=== |
|||
====Start the scan application==== |
|||
*At the System Menu, choose the Scan icon. A blank Scan window appears. The Scan window is divided into areas that display results of scans.The Scan window displays the recipe name in the title bar and the scan sequence in the status box. Scan sequence indicates the order in which slots are scanned: from top-to-bottom or bottom-to-top. |
|||
*Choose the recipe to be used |
|||
*Load the cassette of substrates onto the right locator |
|||
*Choose scan options |
|||
*Enter a Lot ID, if desired |
|||
====Scanning methods==== |
|||
This section describes scanning methods. The procedure describes how to scan all substrates, selected substrates, or one substrate from a cassette(called direct access). The table at the end of this section tells you where to go in this manual to use advanced scan features and options. The recipe sets the primary data of interest (defect or haze) and the initial data displays (map or histogram or both). For each scan, the instrument displays the primary data and lists the results of the scan in the summary. If the recipe specifies alternate data, the data can be manually chosen for display. The recipe also can specify automatic saving or printing of data. When the last substrate in the cassette is scanned or when you press HOME, the instrument homes the cassette and displays the Lot Summary window. (The Lot Summary screen does not appear if Suppress Lot Summary Display is enabled in the User Configuration dialog box.) The Cassette catalog lists the occupied slots of the cassette. The catalog can be numbered from bottom-to-top (the default) or top-to-bottom and the scanning sequence can be set to bottom-to-top or top-to-bottom. |
|||
Scanning options are: automatic or manual operation, scan sequence, and microscope hold for MicroViewing. |
|||
====Using the scan window==== |
|||
When you scan a substrate, the instrument displays the results in the scan summary. Depending upon recipe settings, the instrument also displays scan results in a map, histogram, or both. Data are color-coded as shown by the bin splits in the summary. The cassette catalog displays the contents of the cassette and can be used to select individual substrates for scanning. The Scan window contains the following areas: summary box, wafer map, cassette catalog, histogram, and status box. In the Scan menu bar, choosing ID, Cass, Start, Auto, Home, or Save is the same as pressing the key of the same name on the instrument keypad. Recipe, XY, Print, and Display are pull-down menus. |
|||
<u>The Recipe menu</u> allows you to load a new recipe, view and modify the current recipe, create a new recipe, save a recipe, set user IDs, delete a recipe, enter the Lot ID, extend the puck for inspection and cleaning, and exit the Scan application. For details on recipes, see Chapter 5, “Recipes.” <u>The Print menu</u> allows you to print wafer summaries and lot summaries on the text printer. Summaries can be printed in 40, 80, or 132 column format. The Select Printers item allows you to select the text and graphics printers for the system. The Resume Printer item starts a suspended print job. See Section 4.11, “Printing Summaries,” for details on printing. <u>The Display menu</u> provides data display combinations: defect map only, defect histogram only, defect map plus histogram, haze map only, haze histogram only, or haze map plus histogram. The recipe specifies the initial data display. <u>The Help menu</u> provides help topics. Choose Using Scan to view the help topics related to the Scan application or choose Contents to open the help system. Choosing Using Help displays the Microsoft Windows Using Help topics. |
|||
====Using the summary box==== |
|||
The summary displays the date and time of the scan, the results of the scan, bin splits, overlay controls, and a selection of recipe parameters. The bin splits are color-coded, as shown by the numbered buttons. You can enable or disable a bin from the map or histogram by selecting a bin split button. The summary updates when you enable or disable a bin split, edit a defect or haze bin, or magnify the map. Buttons 1 through 8 control the bin splits for the primary data. Button 0 overlays the alternate data. Button 9 overlays area data. For example, all haze data can be overlayed onto a defect map by selecting button 0. The summary lists primary data and alternate data (if one is selected in the recipe). |
|||
The recipe sets: Primary data of interest (defect or haze); Initial data displays (map or histogram or both). |
|||
To add the comment in the summary screen, press the minus key on the keypad or the minus button (Top left corner) |
|||
=====Data from summary box===== |
|||
*LPD Cnt - the total of all light point defects and their total surface area |
|||
*Bins (1-8) - Bin splits intervals and the count of LPDs for each bin |
|||
*Mean (T) - the mean of collected LPDS |
|||
*Std Dev - the standard deviation of collected LPDs |
|||
*Area Cnt –The count of all areas (areas and scratches) on the substrate and the their total surface area |
|||
*Scratch Cnt-The total number of scratches and their total surface area |
|||
*Sum of All Defects-The sum of all LPDs and areas, including scratches |
|||
*Haze Region-The percentage of the measurable surface area containing haze |
|||
*Haze average- The sum of all haze values divided by the number of haze values, including those that are over the Haze Range limit and under the Haze From limit. When the haze map is zoomed, only those haze values in the magnified map are included in the average, including values under or over the haze range limits. When the haze histogram is zoomed, only those haze values within in the zoomed histogram interval are included in the haze average. Values under or over the haze range limits are not included. |
|||
*Haze peak - The highest haze value |
|||
=====Using the cassette catalog===== |
|||
The Scan application displays the <u>'''Cassette catalog'''</u> at the right of the screen. To display the contents of the cassette loaded on the indexer, press CASS. The Cassette catalog displays the cassette’s substrate status, as follows: |
|||
*If the status of a slot cannot be determined, the slot contains a question mark (?). |
|||
*A white slot number indicates that the substrate has not yet been scanned. |
|||
*A green slot number indicates that the substrate has been scanned and has passed all pass/fail criteria. |
|||
*A red slot number indicates that the substrate has been scanned and has failed one or more pass/fail criteria. |
|||
*A yellow slot number indicates that the instrument is processing the substrate. |
|||
=====Using the histogram===== |
|||
The histogram displays the distribution of light point defects or haze values. The color coding corresponds to the bin splits shown in the summary. Using the histogram window, you can zoom the histogram and change bin splits. A LPD histogram plots the LPD count versus LPD diameter or cross-section as specified in the recipe. A haze histogram plots the number of defects versus haze values. Defect histograms display a red total area bar at the right. |
|||
There are options as: 1) Zoom the histogram (allowing you to narrow the data range); 2) Edit the bin split (the histogram can be used to change LPD bin splits). |
|||
=====Using the wafer mapper===== |
|||
The Scan application can display a wafer map of LPDs or haze. The wafer map can be displayed in combination with a histogram. The recipe sets the initial substrate map/ histogram display combination and the combination can be changed for the current scan by using the Display menu. |
|||
=====Zooming the map===== |
|||
The instrument provides a multilevel zoom feature. When you zoom the map, the instrument displays a panning window and magnifies the map. The menu bar lists the instrument keys that can be used when zooming. |
|||
=====Examining a microview===== |
|||
The Surfscan 6000-series MicroView feature allows you to examine a three dimensional representation of the surface of the substrate. The amplitude axis (Z-axis) is evenly scaled from the smallest to largest amplitudes in the MicroView area. |
|||
You can view the MicroView area from different angles, display the MicroView in color or wireframe format, and use the MicroView cursor to obtain approximate defect amplitude or sizing. Also, you can change the threshold level. A MicroView removes the haze data and sizing information is given in approximate diameter or cross section units. |
|||
===Recipes=== |
|||
Recipes allow you to specify data collection, data display, configuration, pass/fail sort settings, and automatic saving and printing operations for a given wafer substrate size and type. To create a new recipe or modify an existing recipe, you must know how to use Recipe menus and the Recipe dialog box, and know how to set recipe parameters. Also, your user account must allow you to create, edit, and save recipes. The instrument stores recipes in the database by name. A recipe name can be up to 19 characters long and can contain any combination of alphabetic, numeric, or special characters. When you load a recipe from the database, the recipe becomes the current recipe, or the recipe used for scanning. Scan and Setup application windows display the current recipe name in parentheses in their title bars, and the Recipe dialog box displays the current recipe name as its title in parentheses. For step-by-step instructions on setting recipe parameters, see Section 5.3.5, “Selecting Substrate Parameters.” Recipe actions are available from the following applications: |
|||
SCAN or SETUP application - to create, |
|||
DATABASE Export/Import utility - to export or import recipes |
|||
DATABASE Reports utility - to print a recipe listing |
|||
====Recipe menus==== |
|||
Recipe menus in the Scan and Setup applications provide recipe actions: |
|||
*Load - Open and load a selected recipe. Displays the recipe selection dialog box. |
|||
*View/Modify – View or change the current recipe. Displays the Recipe dialog box. |
|||
*New - Create a new recipe. Displays the New Name dialog box. Type the recipe name and choose OK. The instrument sets initial data collection, data display (except haze range and haze from), and configuration values to the previous recipe’s values. The instrument sets haze display and sort parameters to maximum values. Use View/Modify to change recipe settings. |
|||
*Save- save the recipe using the current recipe name |
|||
*Save as- Save the current recipe using a new name. Displays the New Name dialog box |
|||
*Delete- Delete a selected recipe. Displays the selection dialog box. |
|||
====Using the recipe dialog box==== |
|||
A recipe can be created, modified, or viewed by opening the Recipe dialog box from the Scan or Setup applications. The dialog box contains four general parameter areas: data collection, data display, configuration, and sort. |
|||
====Recipe parameters==== |
|||
This section describes the parameters listed in the Recipe dialog box. |
|||
[[File:Recipe parameters.jpg|none|thumb|Recipe Dialog Box|560x560px]] |
|||
======Data collection parameters====== |
|||
1) <u>Substrate diameter</u> (wafers 2-8"); 2) <u>Edge exclusio</u>n (the distance from the edge of the wafer toward the center that is excluded from scan results. Minimum value is 1 mm. Maximum value is one-half wafer diameter); 3) <u>Flat/Notch exclusion</u> (This is the wafer exclusion pattern to apply. One or more exclusion regions can be defined by the exclusion patterns listed in this field. The areas of the wafer specified by exclusion regions are excluded from scan results. See Section 9.5, “Defining Exclusion Patterns,” for details); 4) <u>Haze Suppression</u> (Increases the Haze Range and Haze From by the value entered); 5) <u>Gain/Max Size</u> ( Gain determines the range of data collected during a scan. Gain is a value from 1- 8 for the Surfscan 6200. Max Size indicates the maximum LPD size for this gain. When you change Gain, the instrument automatically sets Area From to Max Size, and sets Threshold and Haze Range to their default values); 6) <u>Threshold</u> (Threshold specifies the smallest LPD size to be included in the scan result. Threshold defaults to the standard value for the current gain setting. If you set threshold to 0, the instrument automatically supplies the default value. If you set threshold to a value equal to or greater than Max Size, data will be collected for one size only); 7) <u>Throughput</u> (The speed at which the wafer is processed through the instrument: high, medium, normal, or low). |
|||
To learn more about Gain/Max size and Threshold values read in [https://wiki.nanotech.ucsb.edu/w/images/9/96/Surfscan-Operation-Manual.pdf Operation-Manual.pdf]. |
|||
======Data display parameters====== |
|||
Data display parameters specify the primary and secondary data, type of display (map, histogram, or both), show size (diameter or cross section), bin splits, area boundary, and haze range. 1) <u>Display option</u> (this is data to collect, display, and save. ''Defect/Haze'': the instrument collects defect and haze data and displays defect data when the scan completes. Haze data can be overlaid by selecting the summary table 0 button. ''Haze/Defect'': the instrument collects defect and haze data and displays haze data when the scan completes. Defect data can be overlaid by selecting the summary table 0 button. ''Defects Only'': the instrument collects, displays, and saves defect data only. ''Haze Only'': the instrument collects, displays, and saves haze data only); 2) <u>Display Type</u> ( shows the initial map and histogram display combination); 3) <u>Show</u> (LPD size calculation and display parameter: diameter (µm) or cross section (µm2 )); 3) <u>Bin Split</u> (Bin split value indicates number of bins used. The Bin Split dialog box allows you to edit LPD bin split ranges and to set the number of bins used); 4) <u>Area From</u> (This value determines the LPD size above which the instrument classifies LPDs as areas. Defaults to the current Max Size value. Maximum diameter is 9900 µm and maximum cross section is 99000 µm2 ); 5) <u>Haze Range</u> (Limited, full, extended or autorange. The haze value range, in parts-per-million, are determined by the current gain setting. In autorange, the instrument chooses the range that provides the best resolution. The instrument automatically zooms on the peak of the data, if any); 6) <u>Haze From</u> (The minimum haze value collected. Defaults to 1/250 of the Haze Range value). |
|||
======Configuration parameters====== |
|||
Configuration parameters provide cassette, sorting, automatic operation, and substrate selection( type and thickness). 1) <u>Cassette</u> (describes cassette type and wafer handling configuration); 2)<u>Sorting</u> (Off); 3) <u>Auto Align [x-y]</u>(Off); 4)<u>Auto Gain Setting</u> (Off); 4) <u>Auto Print</u> (Off); 5) <u>Auto Save</u> (Off); 6) <u>Substrate name</u> (describes calibrated substrate type and thickness. To select a substrate type, choose the SEL button. The dialog box lists the substrate types and thicknesses currently available. Use the cycle control to select an available film thickness. (T) = Tencor-supplied calibration curve (C) = customer-supplied calibration curve). |
|||
======Sort parameters====== |
|||
Establish the pass/fail limits for wafers. If scan results exceed any of the enabled sort parameters in the recipe, the wafer fails, otherwise the wafer passes. Certain recipe parameters interact. When you change Gain, for example, Threshold, Area From, and Haze Range values automatically change to the default values for that gain. You can change the values for these parameters after setting the gain. 1) LPD Count (The maximum number of light particle defects (LPD) allowed: 0 to 99999); 2) <u>LPD/cm2</u> (Number of light particle defects allowed per square centimeter. Defaults to measurable wafer surface area / total LPD); 3) <u>Bin limit</u> (Off or On. Set to On to use the bin limits specified in the Bin Limit dialog box. Bin limits set the number of LPDs allowed for each LPD bin); 4) <u>Area Count</u> (Total number of areas allowed (areas + scratches). Defaults to 9999); 5) <u>Area mm2</u> (Total of all areas allowed in mm2 , 0 to 9999); 6) <u>Scratch Count</u> (Number of scratches allowed, 0 to 9999. Defaults to 9999); 7) <u>Scratch mm</u> (Total of all scratches allowed in mm); 8) <u>Sum of All</u> (The total of LPD Count and Area Count allowed. Defaults to 9999); 9) <u>Haze Average</u> (The maximum allowable haze, when expressed as the average of haze (in ppm) over the measurable surface area); 10) <u>Haze Deviation</u> (The allowable amount of deviation of the haze level from the average, expressed in either absolute (PPM) or relative (percent) terms, over a user-selectable percentage of the wafer’s surface. See Section 5.7.1, “Haze Deviation Sort Parameters,” for details); 11) <u>Haze Peak</u> (The maximum allowable haze value in parts-per-million); 12) <u>Haze Region</u> (The maximum allowable wafer area containing haze, expressed as a percentage of the measurable wafer surface area). |
|||
======Selecting substrate parameters====== |
|||
Setting recipe parameters starts with selecting substrate parameters. A recipe specifies the diameter and edge exclusion for the substrate, and specifies which substrate/film thickness calibration curve to use for calculating LPD sizes. The instrument collects data only from within the measurable surface area of a wafer. |
|||
Choose the select button at the bottom-left of the Recipe dialog box. The dialog box lists all substrates and thicknesses currently calibrated for the instrument. If the substrate you want is not listed, you can calibrate the instrument for the new substrate. See Chapter 7, “Calibration" in [https://wiki.nanotech.ucsb.edu/w/images/9/96/Surfscan-Operation-Manual.pdf Operation-Manual.] |
|||
When a Tencor Instruments-supplied calibration curve is used, the substrate name is followed by (T) in the Recipe dialog box. When a customer-supplied calibration curve is used, the substrate name is followed by (C) in the Recipe dialog box. The (T) or (C) designation also is shown in the wafer summary in the Mean field. To use the recipe limits used when calibrating for the film and thickness specified, check the Recipe Limits from Calibration Curve check box. |
|||
====Setting the data collection range==== |
|||
This section tells you how to obtain the desired data collection range by selecting instrument gain, threshold, the defect-to-area boundary, and haze range for a recipe. |
|||
The instrument collects <u>defect</u> and <u>haze</u> data from a wafer by illuminating the wafer surface with a laser beam, collecting the scattered light through an optics system, and amplifying the scattered light with a photomultiplier tube (PMT). The gain of the PMT determines the dynamic range for collecting data. |
|||
=====Selecting an LPD range===== |
|||
To specify the light point defect (LPD) range, use Gain, Threshold, and Area From parameters. The <u>Gain</u> setting determines the maximum LPD size of interest (the Max Size recipe parameter). <u>Threshold</u> determines the smallest LPD collected during a scan and can be adjusted as necessary to isolate a particular LPD range. The <u>Area From</u> parameter determines the LPD size at which the instrument classifies <u>areas</u>. The instrument classifies LPDs at or above the Area From value as an area. Area From defaults to Max Size (Fig. 5-6) but can be increased if necessary (Fig. 5-7). Using a combination of gain and threshold settings, you can scan for a broad or narrow band of LPDs, depending upon your requirements. The instrument collects, displays, and records LPDs within the range set by the Threshold and Area From parameters. |
|||
*<u>Threshold</u>=Min Size Particles (bin#1) |
|||
*<u>Max Size</u>= Max Size Particles (bin#8) |
|||
*<u>Area From</u> defaults to Max Size |
|||
*<u>Area</u>= determines LPDs above "Area from" |
|||
*True Dynamic Range= LPDs that are collected |
|||
[[File:LPD Ranges.png|thumb|486x486px|none]] |
|||
[[File:Area from Equals Max size.png|thumb|489x489px|none]] |
|||
=====Selecting a Haze range===== |
|||
The haze range collected during a scan is determined by the gain setting, Haze Range value, and Haze From value. Haze Range determines the maximum haze value collected during a scan. You can select from the three haze ranges for each gain setting (full, limited, extended)., or choose the Autorange setting (the instrument chooses the range that provides the best resolution). |
|||
Example: 1) Gain2 [haze range (extended) is set to (39.2–10000)ppm]; 2) Gain4 [haze range (extended) is set to (1.96–500)ppm] |
|||
Haze From is the smallest haze value collected. Haze From defaults to 1/250 the current Haze Range setting but can be set to a different value, as needed. During a scan, the instrument collects haze data from the measurable wafer surface, separated into 832 x 860-µm segments called tiles. Haze maps display the average haze value per tile. Haze histograms display the distribution of haze values. Average haze for a wafer is calculated by averaging the haze values of all tiles on the wafer. |
|||
======What is "Haze"?====== |
|||
Haze is a measure of the light scattered off of a surface. Lower roughness translates into less scattered light and a lower haze. Residual particles scatter light and also increase haze. |
|||
For example, surface roughness, or high-density particulates that are smaller than the tool detection limit could be included as "Haze". |
|||
Typical haze values are less than 0.1 ppm in the narrow normal channel for a highly polished, reflective surface. |
|||
=====Selecting throughput===== |
|||
Throughput (low, normal, medium and high) affects the wafer processing speed, relative instrument sensitivity, and microscan viewing area dimensions. Low throughput results in highest instrument sensitivity,. but produces the lowest wafer-per-hour processing speed. In general, use the highest throughput that produces acceptable results. |
|||
====Selecting display options==== |
|||
This section describes <u>defect</u> and <u>haze</u> display options. The recipe specifies which types of data will be collected during a scan, how to display data in the Scan window (wafer map, histogram, or both), the LPD size calculation method (LPD cross section in µm2 or LPD diameter in µm), defect bin splits, haze bin splits, and initial area and scratch display. |
|||
=====Selecting the data display===== |
|||
The options are: 1) <u>Defect/Haze</u>, 2) Haze/Defect, 3) Defect only, 4) Haze only. |
|||
<u>Defect /Haze</u> (The instrument collects all data and displays defect data in maps and histograms. Haze data can be displayed in the wafer map by using the 0 button in the summary table or haze data can be chosen for display through the Display menu). |
|||
=====Displaying defects===== |
|||
======Light point defects====== |
|||
The instrument color-codes LPD bin splits. You can set the number of LPD bin splits by setting the recipe Bin Split parameter. The following rules apply to setting bin splits: |
|||
*The number of LPD bins can be from one to eight. |
|||
*All bins must be within the data collection range displayed at the bottom of the dialog box. |
|||
*Each bin represents a separate range of values. Bins cannot overlap. |
|||
*The value of the lower boundary of a bin defaults to the value of the upper boundary of the previous bin (For example, if the upper boundary of bin #2 is set to 0.379, the lower boundary of bin #3 is automatically set to 0.379. The lower boundary of bins cannot be set directly). |
|||
*The Area From value defaults to the Max Size value, but can be changed. When Area From exceeds Max Size, the LPD range is extended to the Area From value, and the ranges of LPD bins and bin split limits are adjusted, as necessary. |
|||
======Areas and scratches====== |
|||
The instrument classifies a defect larger than the recipe Area From value as <u>an area</u> or as <u>a scratch</u>. Areas are displayed as red regions on wafer maps. The total surface area occupied by areas is shown by a red vertical bar in histograms. The instrument estimates the size of an area by totaling the length of all chords recorded for the area. The instrument defines a scratch as an area that when an ellipse is drawn around the area. |
|||
• The length of the major axis of the ellipse is equal to or greater than the Scratch Minimum Length value specified in the User Configuration dialog box. |
|||
• The ratio of the major axis of the ellipse to the estimated width must be equal to or greater than the Scratch Length To Width Ratio value specified in the Detailed Scratch Definition dialog box. The instrument estimates width by dividing the area by the length of the major axis of the ellipse. |
|||
• The ratio of the major axis to the minor axis of the ellipse must be equal to or greater than three. The instrument reports scratch count and area total in the wafer summary table. |
|||
======Displaying haze====== |
|||
When the Display Option parameter is set to <u>Haze/Defect</u> or <u>Haze Only</u>, haze is the primary data. The instrument automatically displays haze data in the Scan window for each wafer scan. When Display Option is set to Defect/Haze, haze is the secondary data. The instrument collects haze data which can be displayed by choosing a haze display option from the Display menu. The instrument color-codes haze data. Each color identifies a separate range of data, called a bin split. Haze bin splits can be edited through the Detailed Haze Bin Split dialog box. |
|||
====Selecting Automatic Operations==== |
|||
This section describes the automatic operations available through a recipe, including |
|||
*Automatic sorting of failed wafers to the left cassette |
|||
*Automatic alignment of the wafer to a feature |
|||
*Automatic printing of summaries or the screen |
|||
*Automatic saving of summaries or wafer maps |
|||
====Selecting sort options==== |
|||
Recipe sort parameters determine when a wafer passes or fails the scan. Limits can be set for total number of defects found, defects/cm2 , LPDs per bin, area count, scratch count, area total, scratch length total, haze as a percentage of total measurable wafer area, average haze, and peak haze. A wafer passes when data does not exceed any limits that are On. |
|||
====Recipe parameters guidelines==== |
|||
*Area Total should be less than 10 mm2 for a 150-mm wafer. Adjust range using Gain and Threshold parameters to reduce total area. |
|||
*If the instrument displays Data Overload in the status box, instrument sensitivity is too high. Count accuracy and repeatability might not be reliable. Reduce sensitivity by raising threshold, reducing gain, or increasing throughput. |
|||
*If the histogram shows an exponentially increasing distribution at the low end of the graph, the instrument might be collecting phantom defects or haze noise. Raise threshold and gain proportionately. |
|||
Latest revision as of 00:05, 5 January 2024
Biljana Stamenic, NanoFab Staff, April 2020
Below is the procedure we use for calibrating particle counts on various machines, primarily depositions.
Wafers to Use
The wafers for process calibration are ordered from a wafer supplier that sells high grade, quality wafers. These are 4-inch Si wafers, with low particle count (Particles: <=10 @ >=0.3 microns). Wafers should be handled carefully, because every loading and unloading will add some particulates.
Regular Prime grade wafers also work well, but note that initial pre-process particle measurement may be higher - this is ok as long as you measure before the process step, and subtract that value from the after process value. If your before measurement shows high (eg. >200 particles), do not use that wafer for particle checks, discard it and find a better wafer.
Surfscan 6200 (located in bay5) is used for scanning wafers. There are two standard recipes that could be used for scanning wafers: UCSBTEST1 (for scanning 8” wafers) and UCSBTEST2 (for scanning 4"wafers). The recipe UCSBTEST2 is used as a template to create other two standard recipes: "UCSB Gain4" (measuring small particles 0.16-1.6um) and "UCSB Gain2" (measuring larger particles 1.6-28um).
We have cassettes only for 4", 6" and 8" wafers. If you plan to scan wafers that have different size, you need to provide your own cassette and calibrate it. If you need to scan wafer pieces, you need to develop your own procedure how to scan it.
Scanning procedure for 4" Si wafers
The right-cassette/indexer is calibrated for 4-inch and 6-inch Si wafers. The left-cassette/indexer is available for 8inch Si wafers (not calibrated).
Scan before process step
- Log in (access code is boss)
- Load wafers (face up) to the cassette that says " 4inch Surfscan"
- Place the cassette with wafers to the right indexer
- In the menu option select "CASS" (this will read all wafers that are loaded into the cassette)
- Go to "Recipes"
- Select one of the recipes:
- UCSB G4 0.16-1.6 (measuring small particles 0.16-1.6um)
- UCSB G2 1.6-28.0 (measuring larger particles 1.6-28um)
- Recipe "UCSBTEST2" is used as a template to create recipes UCSB Gain2 and UCSB Gain4
- View the recipe and modify it if needed (if recipe is modified, select bin split/default to save changes)
- Select the wafer in the catalog box you want to scan, and press START (it would scan only one wafer)
- Select AUTO option If you want to scan all wafers in the catalog box, followed by START
- Wait until scan is finished
- Record:
- LPD Cnt (sum of all particles #1- #8), particles #1(0.160-0.213)um, particles #8(1.20-1.60)um
- Haze (region(%), average(ppm), peak(ppm))
- Take a picture of the scan (by phone)
- Select "Home" so indexer moves up to loading/unloading position
- Unload wafers
- Log out
Data Recording
- Run the above process TWICE: once for recipe "Gain 2" (sizes 1.6µm to 28µm) and another for recipe "Gain 4" (sizes 0.160µm to 1.6µm)
- Record the following:
- "total particle count",
- "Haze %" and
- Take photos of the results screen showing all binning and wafer particle distribution. See example photos below.
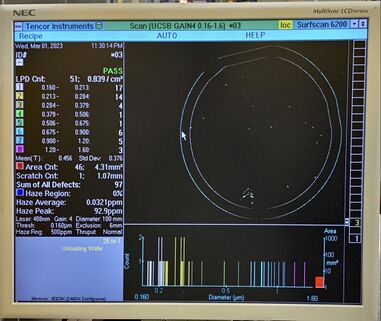
Perform process step
- Run the deposition or other process on your wafer.
- Be careful of adding particles during handling - eg. don't leave wafer out on table unnecessarily, keep it in the case and do not agitate the wafer in the case.
Scan after process calibration
- Repeat the same procedure above for scanning.
- Subtract pre-scan particle counts, to remove pre-process particles that were already on the wafer.
Example Surfscan photos
Low particle example
Here are two example photos for UCSB Gain4, and UCSB Gain2 recipes, showing acceptable particle levels:
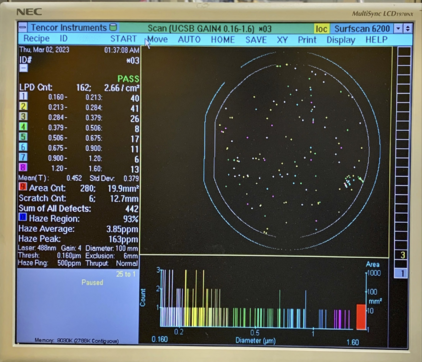
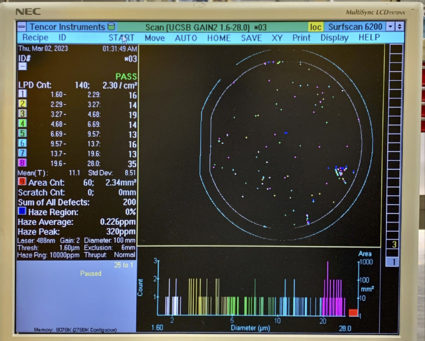
High Particle Example
The following shows examples of high particle levels detected after a deposition process step.
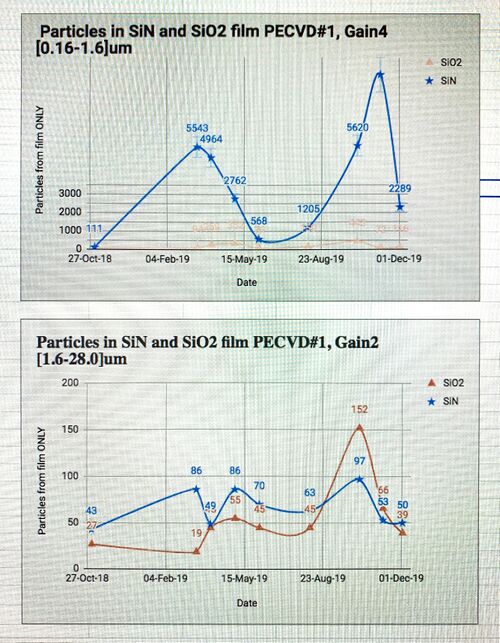
Plotting Process Control Data
Make an Excel spreadsheet. Enter information for Gain4 and Gain2 (particles#1, particles#8, LPD, haze) before and after deposition. For each gain calculate Delta (LPDad - LPDbd). Make a plot (Particulates vs. Date).
Surfscan additional information
Introduction
Surfscan 6200 locates, sizes, and counts defects in semiconductor substrate material down to 0.157um at a 95% capture rate and down to 0.09um at an 80% capture rate. The instrument uses laser beam scanning for detecting defect contamination and displays scan results in color-coded wafer maps, histograms, and summaries. The instrument scans wafers contained in a cassette. Cassettes can be loaded onto the left indexer, right indexer, or both indexers. Cassette configuration can be ( R only, L only, Right to left, Left to right, Sort wafers). When manually aligning wafers, one of the flats of each wafer should face the front of the cassette. When using a mechanical aligner, the flats should face the rear of the cassette. If wafers get scratched, damaged or mishandled immediately perform cassette calibration. The instrument communicates with an operator using the Microsoft Windows. To use an application, choose menus or commands displayed in the menu bar, or use keypad commands. The system menu is an icon-based menu. Double click on the application icon to start an application. In the Scan window , the menu bar displays: ID, CASS, AUTO, HOME, SAVE, and PRINT commands.
Basic skills
This chapter describes instrument controls, including keypad, mouse, floppy drive, and cassette indexer. This chapter also describes how to load a cassette and introduces you to basic program operation using the mouse or keypad.
Getting started
This chapter provides basic startup procedures for the instrument, including: (Powering up the instrument , Logging on to the instrument , Starting Scan , Quitting from an application, Logging off the system, Setting up the system for first use, Using the DOS shell (advanced use), Monitoring disk and database space)
Log in, access code is boss.
Double click on the icon of the application to start application.
Log off the system, to exit the current application.
The Tencor Instrument banner at the top of the screen contains a disk status. If it is green the database space is available. If it is yellow, free disk or data space is getting low, so it needs to be increased free space. It is red, there is no free space.
Scanning
Start the scan application
- At the System Menu, choose the Scan icon. A blank Scan window appears. The Scan window is divided into areas that display results of scans.The Scan window displays the recipe name in the title bar and the scan sequence in the status box. Scan sequence indicates the order in which slots are scanned: from top-to-bottom or bottom-to-top.
- Choose the recipe to be used
- Load the cassette of substrates onto the right locator
- Choose scan options
- Enter a Lot ID, if desired
Scanning methods
This section describes scanning methods. The procedure describes how to scan all substrates, selected substrates, or one substrate from a cassette(called direct access). The table at the end of this section tells you where to go in this manual to use advanced scan features and options. The recipe sets the primary data of interest (defect or haze) and the initial data displays (map or histogram or both). For each scan, the instrument displays the primary data and lists the results of the scan in the summary. If the recipe specifies alternate data, the data can be manually chosen for display. The recipe also can specify automatic saving or printing of data. When the last substrate in the cassette is scanned or when you press HOME, the instrument homes the cassette and displays the Lot Summary window. (The Lot Summary screen does not appear if Suppress Lot Summary Display is enabled in the User Configuration dialog box.) The Cassette catalog lists the occupied slots of the cassette. The catalog can be numbered from bottom-to-top (the default) or top-to-bottom and the scanning sequence can be set to bottom-to-top or top-to-bottom.
Scanning options are: automatic or manual operation, scan sequence, and microscope hold for MicroViewing.
Using the scan window
When you scan a substrate, the instrument displays the results in the scan summary. Depending upon recipe settings, the instrument also displays scan results in a map, histogram, or both. Data are color-coded as shown by the bin splits in the summary. The cassette catalog displays the contents of the cassette and can be used to select individual substrates for scanning. The Scan window contains the following areas: summary box, wafer map, cassette catalog, histogram, and status box. In the Scan menu bar, choosing ID, Cass, Start, Auto, Home, or Save is the same as pressing the key of the same name on the instrument keypad. Recipe, XY, Print, and Display are pull-down menus.
The Recipe menu allows you to load a new recipe, view and modify the current recipe, create a new recipe, save a recipe, set user IDs, delete a recipe, enter the Lot ID, extend the puck for inspection and cleaning, and exit the Scan application. For details on recipes, see Chapter 5, “Recipes.” The Print menu allows you to print wafer summaries and lot summaries on the text printer. Summaries can be printed in 40, 80, or 132 column format. The Select Printers item allows you to select the text and graphics printers for the system. The Resume Printer item starts a suspended print job. See Section 4.11, “Printing Summaries,” for details on printing. The Display menu provides data display combinations: defect map only, defect histogram only, defect map plus histogram, haze map only, haze histogram only, or haze map plus histogram. The recipe specifies the initial data display. The Help menu provides help topics. Choose Using Scan to view the help topics related to the Scan application or choose Contents to open the help system. Choosing Using Help displays the Microsoft Windows Using Help topics.
Using the summary box
The summary displays the date and time of the scan, the results of the scan, bin splits, overlay controls, and a selection of recipe parameters. The bin splits are color-coded, as shown by the numbered buttons. You can enable or disable a bin from the map or histogram by selecting a bin split button. The summary updates when you enable or disable a bin split, edit a defect or haze bin, or magnify the map. Buttons 1 through 8 control the bin splits for the primary data. Button 0 overlays the alternate data. Button 9 overlays area data. For example, all haze data can be overlayed onto a defect map by selecting button 0. The summary lists primary data and alternate data (if one is selected in the recipe).
The recipe sets: Primary data of interest (defect or haze); Initial data displays (map or histogram or both).
To add the comment in the summary screen, press the minus key on the keypad or the minus button (Top left corner)
Data from summary box
- LPD Cnt - the total of all light point defects and their total surface area
- Bins (1-8) - Bin splits intervals and the count of LPDs for each bin
- Mean (T) - the mean of collected LPDS
- Std Dev - the standard deviation of collected LPDs
- Area Cnt –The count of all areas (areas and scratches) on the substrate and the their total surface area
- Scratch Cnt-The total number of scratches and their total surface area
- Sum of All Defects-The sum of all LPDs and areas, including scratches
- Haze Region-The percentage of the measurable surface area containing haze
- Haze average- The sum of all haze values divided by the number of haze values, including those that are over the Haze Range limit and under the Haze From limit. When the haze map is zoomed, only those haze values in the magnified map are included in the average, including values under or over the haze range limits. When the haze histogram is zoomed, only those haze values within in the zoomed histogram interval are included in the haze average. Values under or over the haze range limits are not included.
- Haze peak - The highest haze value
Using the cassette catalog
The Scan application displays the Cassette catalog at the right of the screen. To display the contents of the cassette loaded on the indexer, press CASS. The Cassette catalog displays the cassette’s substrate status, as follows:
- If the status of a slot cannot be determined, the slot contains a question mark (?).
- A white slot number indicates that the substrate has not yet been scanned.
- A green slot number indicates that the substrate has been scanned and has passed all pass/fail criteria.
- A red slot number indicates that the substrate has been scanned and has failed one or more pass/fail criteria.
- A yellow slot number indicates that the instrument is processing the substrate.
Using the histogram
The histogram displays the distribution of light point defects or haze values. The color coding corresponds to the bin splits shown in the summary. Using the histogram window, you can zoom the histogram and change bin splits. A LPD histogram plots the LPD count versus LPD diameter or cross-section as specified in the recipe. A haze histogram plots the number of defects versus haze values. Defect histograms display a red total area bar at the right.
There are options as: 1) Zoom the histogram (allowing you to narrow the data range); 2) Edit the bin split (the histogram can be used to change LPD bin splits).
Using the wafer mapper
The Scan application can display a wafer map of LPDs or haze. The wafer map can be displayed in combination with a histogram. The recipe sets the initial substrate map/ histogram display combination and the combination can be changed for the current scan by using the Display menu.
Zooming the map
The instrument provides a multilevel zoom feature. When you zoom the map, the instrument displays a panning window and magnifies the map. The menu bar lists the instrument keys that can be used when zooming.
Examining a microview
The Surfscan 6000-series MicroView feature allows you to examine a three dimensional representation of the surface of the substrate. The amplitude axis (Z-axis) is evenly scaled from the smallest to largest amplitudes in the MicroView area.
You can view the MicroView area from different angles, display the MicroView in color or wireframe format, and use the MicroView cursor to obtain approximate defect amplitude or sizing. Also, you can change the threshold level. A MicroView removes the haze data and sizing information is given in approximate diameter or cross section units.
Recipes
Recipes allow you to specify data collection, data display, configuration, pass/fail sort settings, and automatic saving and printing operations for a given wafer substrate size and type. To create a new recipe or modify an existing recipe, you must know how to use Recipe menus and the Recipe dialog box, and know how to set recipe parameters. Also, your user account must allow you to create, edit, and save recipes. The instrument stores recipes in the database by name. A recipe name can be up to 19 characters long and can contain any combination of alphabetic, numeric, or special characters. When you load a recipe from the database, the recipe becomes the current recipe, or the recipe used for scanning. Scan and Setup application windows display the current recipe name in parentheses in their title bars, and the Recipe dialog box displays the current recipe name as its title in parentheses. For step-by-step instructions on setting recipe parameters, see Section 5.3.5, “Selecting Substrate Parameters.” Recipe actions are available from the following applications:
SCAN or SETUP application - to create,
DATABASE Export/Import utility - to export or import recipes
DATABASE Reports utility - to print a recipe listing
Recipe menus in the Scan and Setup applications provide recipe actions:
- Load - Open and load a selected recipe. Displays the recipe selection dialog box.
- View/Modify – View or change the current recipe. Displays the Recipe dialog box.
- New - Create a new recipe. Displays the New Name dialog box. Type the recipe name and choose OK. The instrument sets initial data collection, data display (except haze range and haze from), and configuration values to the previous recipe’s values. The instrument sets haze display and sort parameters to maximum values. Use View/Modify to change recipe settings.
- Save- save the recipe using the current recipe name
- Save as- Save the current recipe using a new name. Displays the New Name dialog box
- Delete- Delete a selected recipe. Displays the selection dialog box.
Using the recipe dialog box
A recipe can be created, modified, or viewed by opening the Recipe dialog box from the Scan or Setup applications. The dialog box contains four general parameter areas: data collection, data display, configuration, and sort.
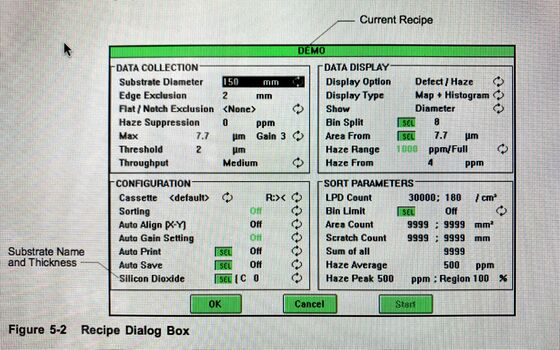
Recipe parameters
This section describes the parameters listed in the Recipe dialog box.
Data collection parameters
1) Substrate diameter (wafers 2-8"); 2) Edge exclusion (the distance from the edge of the wafer toward the center that is excluded from scan results. Minimum value is 1 mm. Maximum value is one-half wafer diameter); 3) Flat/Notch exclusion (This is the wafer exclusion pattern to apply. One or more exclusion regions can be defined by the exclusion patterns listed in this field. The areas of the wafer specified by exclusion regions are excluded from scan results. See Section 9.5, “Defining Exclusion Patterns,” for details); 4) Haze Suppression (Increases the Haze Range and Haze From by the value entered); 5) Gain/Max Size ( Gain determines the range of data collected during a scan. Gain is a value from 1- 8 for the Surfscan 6200. Max Size indicates the maximum LPD size for this gain. When you change Gain, the instrument automatically sets Area From to Max Size, and sets Threshold and Haze Range to their default values); 6) Threshold (Threshold specifies the smallest LPD size to be included in the scan result. Threshold defaults to the standard value for the current gain setting. If you set threshold to 0, the instrument automatically supplies the default value. If you set threshold to a value equal to or greater than Max Size, data will be collected for one size only); 7) Throughput (The speed at which the wafer is processed through the instrument: high, medium, normal, or low).
To learn more about Gain/Max size and Threshold values read in Operation-Manual.pdf.
Data display parameters
Data display parameters specify the primary and secondary data, type of display (map, histogram, or both), show size (diameter or cross section), bin splits, area boundary, and haze range. 1) Display option (this is data to collect, display, and save. Defect/Haze: the instrument collects defect and haze data and displays defect data when the scan completes. Haze data can be overlaid by selecting the summary table 0 button. Haze/Defect: the instrument collects defect and haze data and displays haze data when the scan completes. Defect data can be overlaid by selecting the summary table 0 button. Defects Only: the instrument collects, displays, and saves defect data only. Haze Only: the instrument collects, displays, and saves haze data only); 2) Display Type ( shows the initial map and histogram display combination); 3) Show (LPD size calculation and display parameter: diameter (µm) or cross section (µm2 )); 3) Bin Split (Bin split value indicates number of bins used. The Bin Split dialog box allows you to edit LPD bin split ranges and to set the number of bins used); 4) Area From (This value determines the LPD size above which the instrument classifies LPDs as areas. Defaults to the current Max Size value. Maximum diameter is 9900 µm and maximum cross section is 99000 µm2 ); 5) Haze Range (Limited, full, extended or autorange. The haze value range, in parts-per-million, are determined by the current gain setting. In autorange, the instrument chooses the range that provides the best resolution. The instrument automatically zooms on the peak of the data, if any); 6) Haze From (The minimum haze value collected. Defaults to 1/250 of the Haze Range value).
Configuration parameters
Configuration parameters provide cassette, sorting, automatic operation, and substrate selection( type and thickness). 1) Cassette (describes cassette type and wafer handling configuration); 2)Sorting (Off); 3) Auto Align [x-y](Off); 4)Auto Gain Setting (Off); 4) Auto Print (Off); 5) Auto Save (Off); 6) Substrate name (describes calibrated substrate type and thickness. To select a substrate type, choose the SEL button. The dialog box lists the substrate types and thicknesses currently available. Use the cycle control to select an available film thickness. (T) = Tencor-supplied calibration curve (C) = customer-supplied calibration curve).
Sort parameters
Establish the pass/fail limits for wafers. If scan results exceed any of the enabled sort parameters in the recipe, the wafer fails, otherwise the wafer passes. Certain recipe parameters interact. When you change Gain, for example, Threshold, Area From, and Haze Range values automatically change to the default values for that gain. You can change the values for these parameters after setting the gain. 1) LPD Count (The maximum number of light particle defects (LPD) allowed: 0 to 99999); 2) LPD/cm2 (Number of light particle defects allowed per square centimeter. Defaults to measurable wafer surface area / total LPD); 3) Bin limit (Off or On. Set to On to use the bin limits specified in the Bin Limit dialog box. Bin limits set the number of LPDs allowed for each LPD bin); 4) Area Count (Total number of areas allowed (areas + scratches). Defaults to 9999); 5) Area mm2 (Total of all areas allowed in mm2 , 0 to 9999); 6) Scratch Count (Number of scratches allowed, 0 to 9999. Defaults to 9999); 7) Scratch mm (Total of all scratches allowed in mm); 8) Sum of All (The total of LPD Count and Area Count allowed. Defaults to 9999); 9) Haze Average (The maximum allowable haze, when expressed as the average of haze (in ppm) over the measurable surface area); 10) Haze Deviation (The allowable amount of deviation of the haze level from the average, expressed in either absolute (PPM) or relative (percent) terms, over a user-selectable percentage of the wafer’s surface. See Section 5.7.1, “Haze Deviation Sort Parameters,” for details); 11) Haze Peak (The maximum allowable haze value in parts-per-million); 12) Haze Region (The maximum allowable wafer area containing haze, expressed as a percentage of the measurable wafer surface area).
Selecting substrate parameters
Setting recipe parameters starts with selecting substrate parameters. A recipe specifies the diameter and edge exclusion for the substrate, and specifies which substrate/film thickness calibration curve to use for calculating LPD sizes. The instrument collects data only from within the measurable surface area of a wafer.
Choose the select button at the bottom-left of the Recipe dialog box. The dialog box lists all substrates and thicknesses currently calibrated for the instrument. If the substrate you want is not listed, you can calibrate the instrument for the new substrate. See Chapter 7, “Calibration" in Operation-Manual.
When a Tencor Instruments-supplied calibration curve is used, the substrate name is followed by (T) in the Recipe dialog box. When a customer-supplied calibration curve is used, the substrate name is followed by (C) in the Recipe dialog box. The (T) or (C) designation also is shown in the wafer summary in the Mean field. To use the recipe limits used when calibrating for the film and thickness specified, check the Recipe Limits from Calibration Curve check box.
Setting the data collection range
This section tells you how to obtain the desired data collection range by selecting instrument gain, threshold, the defect-to-area boundary, and haze range for a recipe.
The instrument collects defect and haze data from a wafer by illuminating the wafer surface with a laser beam, collecting the scattered light through an optics system, and amplifying the scattered light with a photomultiplier tube (PMT). The gain of the PMT determines the dynamic range for collecting data.
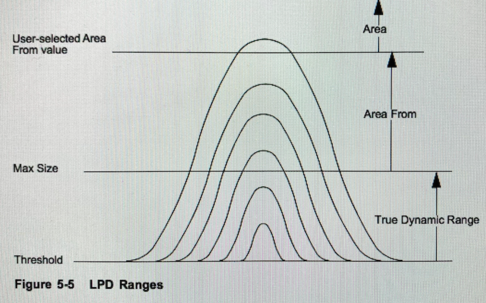
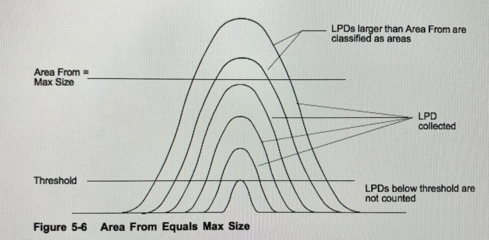
Selecting an LPD range
To specify the light point defect (LPD) range, use Gain, Threshold, and Area From parameters. The Gain setting determines the maximum LPD size of interest (the Max Size recipe parameter). Threshold determines the smallest LPD collected during a scan and can be adjusted as necessary to isolate a particular LPD range. The Area From parameter determines the LPD size at which the instrument classifies areas. The instrument classifies LPDs at or above the Area From value as an area. Area From defaults to Max Size (Fig. 5-6) but can be increased if necessary (Fig. 5-7). Using a combination of gain and threshold settings, you can scan for a broad or narrow band of LPDs, depending upon your requirements. The instrument collects, displays, and records LPDs within the range set by the Threshold and Area From parameters.
- Threshold=Min Size Particles (bin#1)
- Max Size= Max Size Particles (bin#8)
- Area From defaults to Max Size
- Area= determines LPDs above "Area from"
- True Dynamic Range= LPDs that are collected
Selecting a Haze range
The haze range collected during a scan is determined by the gain setting, Haze Range value, and Haze From value. Haze Range determines the maximum haze value collected during a scan. You can select from the three haze ranges for each gain setting (full, limited, extended)., or choose the Autorange setting (the instrument chooses the range that provides the best resolution).
Example: 1) Gain2 [haze range (extended) is set to (39.2–10000)ppm]; 2) Gain4 [haze range (extended) is set to (1.96–500)ppm]
Haze From is the smallest haze value collected. Haze From defaults to 1/250 the current Haze Range setting but can be set to a different value, as needed. During a scan, the instrument collects haze data from the measurable wafer surface, separated into 832 x 860-µm segments called tiles. Haze maps display the average haze value per tile. Haze histograms display the distribution of haze values. Average haze for a wafer is calculated by averaging the haze values of all tiles on the wafer.
What is "Haze"?
Haze is a measure of the light scattered off of a surface. Lower roughness translates into less scattered light and a lower haze. Residual particles scatter light and also increase haze.
For example, surface roughness, or high-density particulates that are smaller than the tool detection limit could be included as "Haze".
Typical haze values are less than 0.1 ppm in the narrow normal channel for a highly polished, reflective surface.
Selecting throughput
Throughput (low, normal, medium and high) affects the wafer processing speed, relative instrument sensitivity, and microscan viewing area dimensions. Low throughput results in highest instrument sensitivity,. but produces the lowest wafer-per-hour processing speed. In general, use the highest throughput that produces acceptable results.
Selecting display options
This section describes defect and haze display options. The recipe specifies which types of data will be collected during a scan, how to display data in the Scan window (wafer map, histogram, or both), the LPD size calculation method (LPD cross section in µm2 or LPD diameter in µm), defect bin splits, haze bin splits, and initial area and scratch display.
Selecting the data display
The options are: 1) Defect/Haze, 2) Haze/Defect, 3) Defect only, 4) Haze only.
Defect /Haze (The instrument collects all data and displays defect data in maps and histograms. Haze data can be displayed in the wafer map by using the 0 button in the summary table or haze data can be chosen for display through the Display menu).
Displaying defects
Light point defects
The instrument color-codes LPD bin splits. You can set the number of LPD bin splits by setting the recipe Bin Split parameter. The following rules apply to setting bin splits:
- The number of LPD bins can be from one to eight.
- All bins must be within the data collection range displayed at the bottom of the dialog box.
- Each bin represents a separate range of values. Bins cannot overlap.
- The value of the lower boundary of a bin defaults to the value of the upper boundary of the previous bin (For example, if the upper boundary of bin #2 is set to 0.379, the lower boundary of bin #3 is automatically set to 0.379. The lower boundary of bins cannot be set directly).
- The Area From value defaults to the Max Size value, but can be changed. When Area From exceeds Max Size, the LPD range is extended to the Area From value, and the ranges of LPD bins and bin split limits are adjusted, as necessary.
Areas and scratches
The instrument classifies a defect larger than the recipe Area From value as an area or as a scratch. Areas are displayed as red regions on wafer maps. The total surface area occupied by areas is shown by a red vertical bar in histograms. The instrument estimates the size of an area by totaling the length of all chords recorded for the area. The instrument defines a scratch as an area that when an ellipse is drawn around the area.
• The length of the major axis of the ellipse is equal to or greater than the Scratch Minimum Length value specified in the User Configuration dialog box.
• The ratio of the major axis of the ellipse to the estimated width must be equal to or greater than the Scratch Length To Width Ratio value specified in the Detailed Scratch Definition dialog box. The instrument estimates width by dividing the area by the length of the major axis of the ellipse.
• The ratio of the major axis to the minor axis of the ellipse must be equal to or greater than three. The instrument reports scratch count and area total in the wafer summary table.
Displaying haze
When the Display Option parameter is set to Haze/Defect or Haze Only, haze is the primary data. The instrument automatically displays haze data in the Scan window for each wafer scan. When Display Option is set to Defect/Haze, haze is the secondary data. The instrument collects haze data which can be displayed by choosing a haze display option from the Display menu. The instrument color-codes haze data. Each color identifies a separate range of data, called a bin split. Haze bin splits can be edited through the Detailed Haze Bin Split dialog box.
Selecting Automatic Operations
This section describes the automatic operations available through a recipe, including
- Automatic sorting of failed wafers to the left cassette
- Automatic alignment of the wafer to a feature
- Automatic printing of summaries or the screen
- Automatic saving of summaries or wafer maps
Selecting sort options
Recipe sort parameters determine when a wafer passes or fails the scan. Limits can be set for total number of defects found, defects/cm2 , LPDs per bin, area count, scratch count, area total, scratch length total, haze as a percentage of total measurable wafer area, average haze, and peak haze. A wafer passes when data does not exceed any limits that are On.
Recipe parameters guidelines
- Area Total should be less than 10 mm2 for a 150-mm wafer. Adjust range using Gain and Threshold parameters to reduce total area.
- If the instrument displays Data Overload in the status box, instrument sensitivity is too high. Count accuracy and repeatability might not be reliable. Reduce sensitivity by raising threshold, reducing gain, or increasing throughput.
- If the histogram shows an exponentially increasing distribution at the low end of the graph, the instrument might be collecting phantom defects or haze noise. Raise threshold and gain proportionately.